Introduction
This notebook shows how to customize the different parts of the BERTopic pipeline: Embeddings, UMAP and HDBSCAN. See this Explainer and tutorial by pinecone.io ) for details.
Imports and Login
import pandas as pdimport numpy as npfrom tqdm.auto import tqdmimport jsonfrom bertopic import BERTopicfrom sentence_transformers import SentenceTransformerimport matplotlib.pyplot as pltimport seaborn as snsimport timeimport torchimport umapimport hdbscan
Load publication data
= pd.read_csv(INPUT_FILE)= df.drop_duplicates(subset= ["doi" ])\ = ["doi" , "abstract" ]).reset_index(drop= True )= (df["title" ] + " " + df["abstract" ]).values
Embeddings
Compute or load sentence embeddings
if COMPUTE_EMBEDDINGS:= SentenceTransformer('allenai-specter' )= embedding_model.encode(sentences, normalize_embeddings= True )else := np.load(OUTPUT_FILE_TOPICS_EMBEDDINGS)
Sample embeddings
= torch.randperm(embeddings.shape[0 ])= perm[:3000 ]= embeddings[idx, :]
UMAP
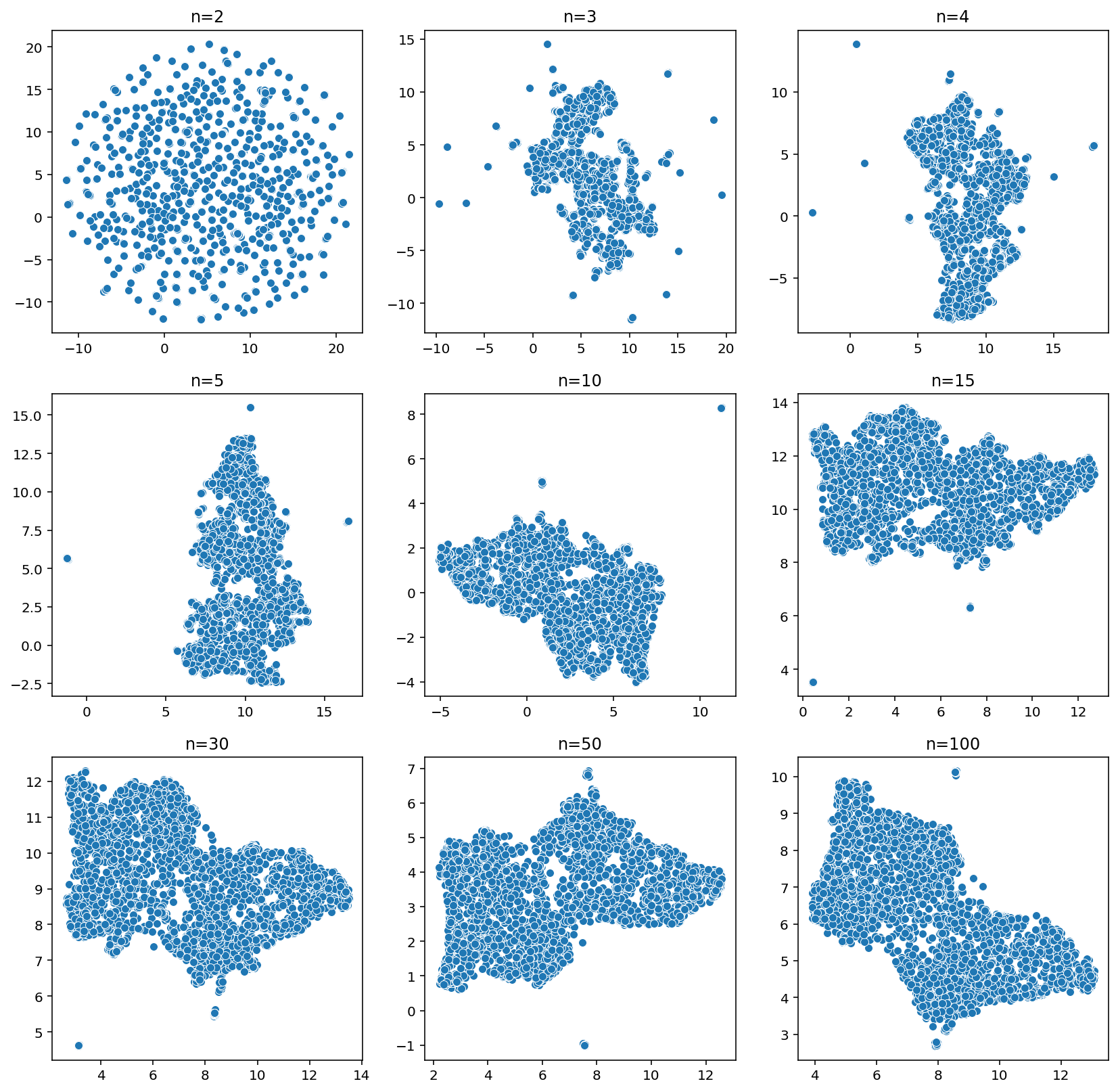
Hyperparameter search for n_neighbors
= plt.subplots(3 , 3 , figsize= (14 , 14 ))= [2 , 3 , 4 , 5 , 10 , 15 , 30 , 50 , 100 ]= 0 , 0 for n_neighbors in tqdm(nns):= umap.UMAP(n_neighbors= n_neighbors)= fit.fit_transform(samples)= u[:,0 ], y= u[:,1 ], ax= ax[j, i])f'n= { n_neighbors} ' )if i < 2 : i += 1 else : i = 0 ; j += 1
OMP: Info #276: omp_set_nested routine deprecated, please use omp_set_max_active_levels instead.
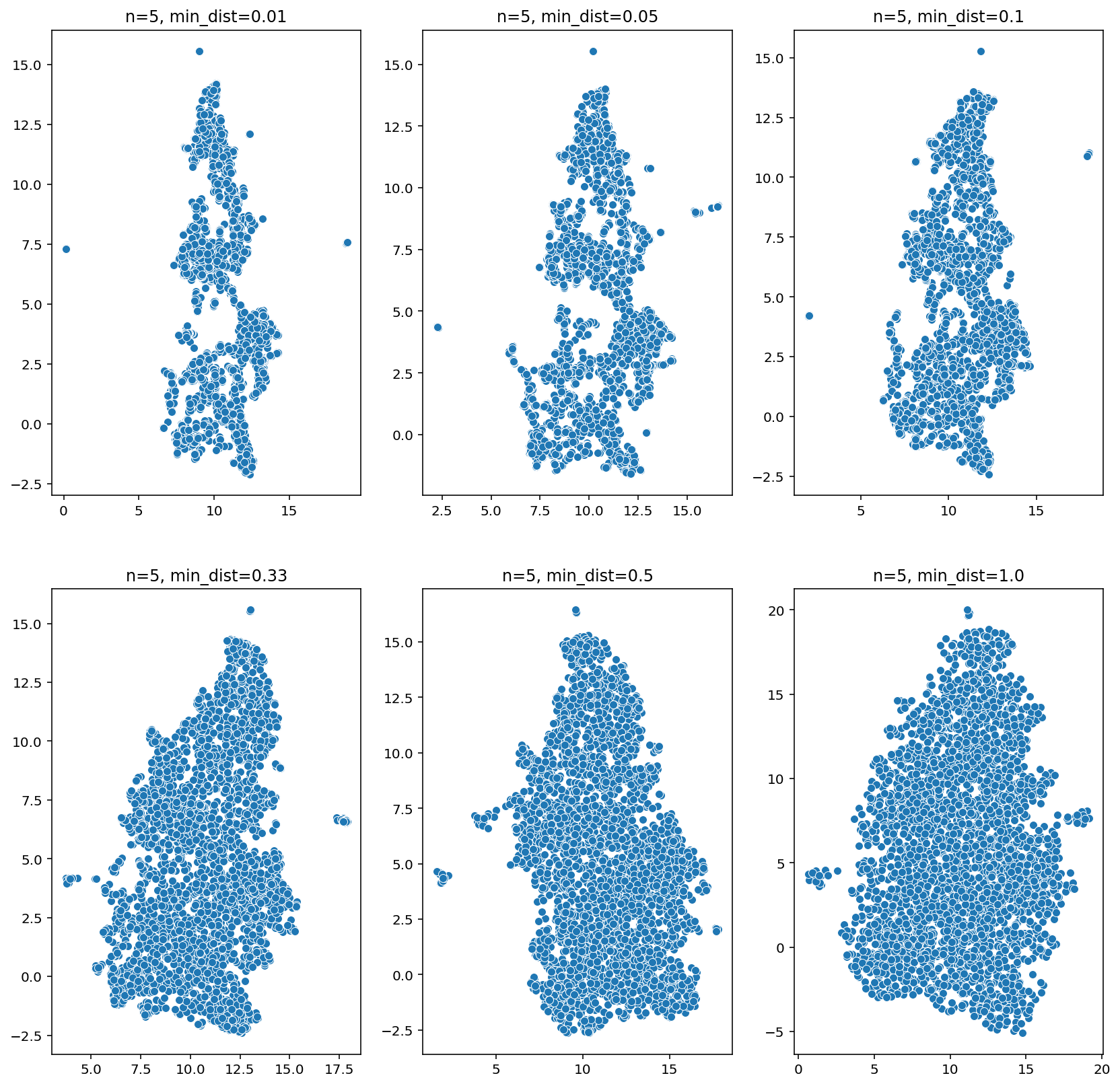
Hyperparameter search for min_dist
= 5 = plt.subplots(2 , 3 , figsize= (14 , 14 ))= [0.01 , 0.05 , 0.10 , 0.33 , 0.5 , 1.0 ]= 0 , 0 for min_dist in tqdm(min_dists):= umap.UMAP(n_neighbors= n_neighbors, min_dist= min_dist)= fit.fit_transform(samples)= u[:,0 ], y= u[:,1 ], ax= ax[j, i])f'n= { n_neighbors} , min_dist= { min_dist} ' )if i < 2 : i += 1 else : i = 0 ; j += 1
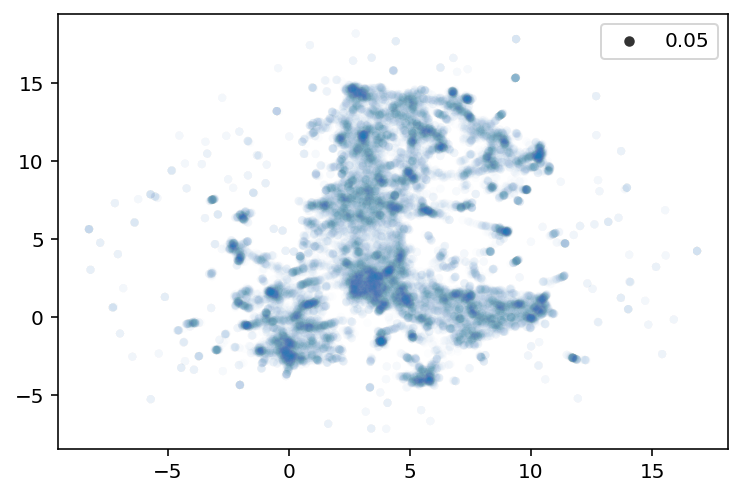
Fit UMAP
= 0.1 = 5 = umap.UMAP(n_neighbors= n_neighbors, min_dist= min_dist)= umap_fitter.fit_transform(embeddings)= u[:,0 ], y= u[:,1 ], alpha= 0.01 , size= 0.05 )
HDBSCAN
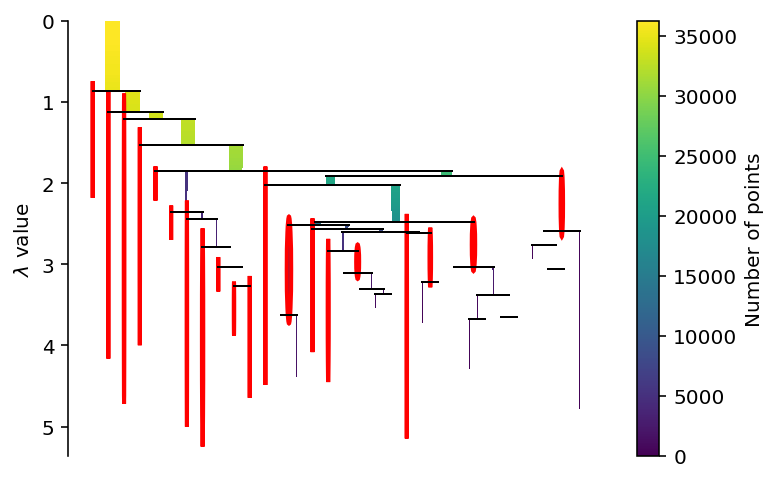
Clustering with defaults
= hdbscan.HDBSCAN()= True )
<AxesSubplot:ylabel='$\\lambda$ value'>
Clustering with optimized parameters
= hdbscan.HDBSCAN(= 200 ,= 100 ,= True ,= True = True )
<AxesSubplot:ylabel='$\\lambda$ value'>
Custom BERTopic Model
BERTopic with customized models
import nltkfrom nltk.corpus import stopwordsfrom sklearn.feature_extraction.text import CountVectorizer'stopwords' )= list (stopwords.words('english' )) + ["covid" , "covid 19" , "19" , "sars" , "cov" , "sars cov" , "patients" , "epidemic" , "pandemic" , "health" , "sec" , "sec sec" ]= CountVectorizer(ngram_range= (1 , 2 ), stop_words= stopwords)if COMPUTE_TOPICS:= BERTopic(= umap_fitter,= clusterer,= embedding_model,= vectorizer_model,= True ,= 5 ,= 'english' ,= True = topic_model.fit_transform(sentences)= False )else := BERTopic.load(OUTPUT_FILE_TOPICS_BERTOPIC)
Visualize topics
= 50 )